Disclaimer: Early release articles are not considered as final versions. Any changes will be reflected in the online version in the month the article is officially released.
Author affiliations: Shanxi Provincial People’s Hospital, Taiyuan, China (Q. Cui); Renji Hospital at Shanghai Jiao Tong University School of Medicine, Shanghai, China (C. Wang, Q. Wang, J. Qin, M. Li, Z. Shen); Huashan Hospital at Fudan University, Shanghai (B. Ding); Key Laboratory of Clinical Pharmacology of Antibiotics, Ministry of Health, Shanghai (B. Ding)
Emergence and global dissemination of carbapenem-resistant Klebsiella pneumoniae pose therapeutic challenges to public health (1). The most crucial cause of carbapenem resistance in K. pneumoniae is carbapenemase production; thus, the novel β-lactamase inhibitor ceftazidime/avibactam (CAZ/AVI) provides an antimicrobial strategy (1–3). However, its increasing use raises resistance concerns. According to the China Antimicrobial Surveillance Network (http://www.chinets.com/Data/AntibioticdrugFast), 9.9% of K. pneumoniae carbapenemase (KPC) 2–producing K. pneumoniae (KPC-KP) displayed CAZ/AVI resistance (4). β-lactamase amino acid substitutions are the dominant mechanisms that lead to CAZ/AVI resistance (5). Mutations in class A β-lactamases, especially KPCs, have been reported (5). Substitutions in KPCs could improve ceftazidime affinity or reduce avibactam inhibition (5). We report a novel CAZ/AVI resistance mechanism in epidemic sequence type (ST) 11 KPC-KP. All study procedures involving human participants and animals were in accordance with the ethics standards of the Institutional Review Board Ethics Committee of Shanxi Provincial People’s Hospital; this type of retrospective study did not require formal consent.
In 2021, a 62-year-old man was transferred from another hospital to a teaching hospital in Shanxi Province, China. Before transfer, a blood culture indicated CAZ/AVI–susceptible carbapenem-resistant K. pneumoniae. The patient received 1 week of CAZ-AVI therapy before transfer and another week of CAZ/AVI therapy after admission. In addition to the bloodstream infection, severe pneumoniae, multiple duodenal ulcers, and gastrointestinal hemorrhage developed. Two weeks after CAZ/AVI withdrawal, we isolated KP0714, which was resistant to all β-lactams tested but susceptible to tigecycline and polymyxin B (Appendix Table 1). We generated the KP0714 complete genome by using the combination of Illumina and PacBio RS sequencing (Appendix Table 2), and it belonged to ST11. The resistance plasmid pKPC-KP0714 carries blaKPC-2 and several other resistance genes, including blaTEM-1, rmtB, and fosA3 (Appendix Table 2), and KPC-2 S130A substitution was constructed in situ. KP0714 and the KPC-2 S130A mutant displayed the same MICs for CAZ/AVI, suggesting that blaKPC-2 was not involved in CAZ/AVI resistance (Appendix Table 1).
KP0714 possessed a novel IncFIB(K)-type virulence plasmid pVir-KP0714, encoding siderophore aerobactin (iucABCDiutA) and capsular polysaccharide regulator RmpA2. pVir-KP0714 was 99.97% identical to reference plasmid pOXA1_020030 (GenBank accession no. CP028791) from K. pneumoniae strain WCHKP020030 at 74% coverage. Both ends of pVir-KP0714 were absent from pOXA1_020030 but were highly homologous to another plasmid, pLAP2_020030 (GenBank accession no. CP028792), from WCHKP020030 (Figure 1; Appendix Figure 1). Multiple mobile genetic elements on these plasmids suggested that pVir-KP0714 was generated through genetic recombination between pOXA1_020030 and pLAP2_020030, which not only formed a novel virulence plasmid but also contributed to chromosomal integration of a 45-kb plasmid fragment from pLAP2_020030 (Figure 1; Appendix Figure 1). The 45-kb fragment that had not been integrated into pVir-KP0714 was divided into the upstream 30-kb fragment and a 15-kb genetic context containing blaSHV-12, which were independently inserted into the chromosome. The 15-kb genetic context containing blaSHV-12 was flanked by several IS26 insertion sequences and harbored 3 other resistance genes, blaLAP-2, qnrS1, and aph(3′)-Ia, which exhibited 100% identity and 100% query coverage with the reference plasmid pLAP2_020030 (Figure 2; Appendix Figure 2).
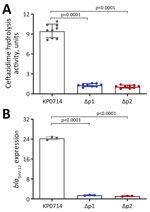
However, we observed substantial structural changes in this chromosomal insertion fragment compared with pLAP2_020030 (Figure 2; Appendix Figure 2). The reversion and rearrangement of IS26-aph(3′)-Ia provided an addition promoter P2 for blaSHV-12 (Figure 2; Appendix Figure 2). To determine the role of promoter P2 in CAZ/AVI resistance, we deleted P2 and the original promoter P1 of blaSHV-12 by using a pConj working vector-based genetic engineering approach (6). Deletion of P1 or P2 could completely restore KP0714 susceptibility to CAZ/AVI; CAZ/AVI MICs were 2 and 1 μg/mL, respectively (Appendix Table 1). The relative expression of blaSHV-12 in KP0714 was ≈20-fold higher than in ΔP1 and ΔP2 mutants (Figure 2; Appendix Figure 2). Similarly, the hydrolysis activity of ceftazidime in KP0714 was significantly higher than that of ΔP1 and ΔP2 mutants (p<0.0001). Those results demonstrated that CAZ/AVI resistance in KP0714 was attributed to overexpression of blaSHV-12 resulting from an additional promoter, and the original promoter P1 was also necessary for the biological function of P2.
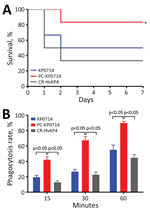
Because a novel virulence plasmid pVir-KP0714 was formed through plasmid recombination, we determined the virulence potential of KP0714 by using a mouse infection model and human neutrophil phagocytosis assay (7). As the hypervirulence control, we used the previously reported ST11 carbapenem-resistant hypervirulent K. pneumoniae strain CR-HvKP4 (8). We found no statistical difference regarding mouse survival and neutrophil phagocytosis between KP0714 and CR-HvKP4 (Figure 1; Appendix Figure 2), suggesting convergence of hypervirulence and CAZ/AVI resistance in KP0714. In contrast, mouse survival rates were significantly higher and human neutrophil phagocytosis rates were significantly lower for KP0714 and CR-HvKP4 at each time point when compared with virulence plasmid pVir-KP0714-curing KP0714 (PC-KP0714), demonstrating that KP0714 hypervirulence was attributed to acquisition of virulence plasmid pVir-KP0714.
In conclusion, KP0714 high-level resistance to carbapenems and CAZ/AVI, compensating for decreased carbapenem hydrolyzation activity of KPC variants (5,9), highlights a novel evolution pathway for development of CAZ/AVI resistance in epidemic ST11 KPC-KP, posing a threat to clinical antimicrobial therapy. Emerging CAZ/AVI-resistant and hypervirulent ST11 KPC-KP might be continuously evolving and warrants prospective monitoring.
Mrs. Cui is a researcher at Shanxi Provincial People’s Hospital of Shanxi Medical University. Her research interests are epidemiology and antimicrobial-resistance mechanisms of carbapenem-resistant Enterobacteriaceae.
We thank the authority of CR-HvKP4 by Rong Zhang from the Second Affiliated Hospital of Zhejiang University.
This study was supported by National Natural Science Foundation of China (82272374), Shanghai Pujiang Program (22PJ1409600), and a research fund from Renji Hospital for young scholars (RJTJ22-MS-018). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The complete genome sequences of KP0714 were deposited in the GenBank (accession nos. CP128191–5).